Asthma is a chronic inflammatory disease of the airways. Asthma patients may experience potentially life-threatening episodic flare-ups, known as exacerbations, which may significantly contribute to the asthma burden. The Pi*S and Pi*Z variants of the SERPINA1 gene, which usually involve alpha-1 antitrypsin (AAT) deficiency, had previously been associated with asthma. The link between AAT deficiency and asthma might be represented by the elastase/antielastase imbalance. However, their role in asthma exacerbations remains unknown. Our objective was to assess whether SERPINA1 genetic variants and reduced AAT protein levels are associated with asthma exacerbations.
Materials and methodsIn the discovery analysis, SERPINA1 Pi*S and Pi*Z variants and serum AAT levels were analyzed in 369 subjects from La Palma (Canary Islands, Spain). As replication, genomic data from two studies focused on 525 Spaniards and publicly available data from UK Biobank, FinnGen, and GWAS Catalog (Open Targets Genetics) were analyzed. The associations between SERPINA1 Pi*S and Pi*Z variants and AAT deficiency with asthma exacerbations were analyzed with logistic regression models, including age, sex, and genotype principal components as covariates.
ResultsIn the discovery, a significant association with asthma exacerbations was found for both Pi*S (odds ratio [OR]=2.38, 95% confidence interval [CI]= 1.40–4.04, p-value=0.001) and Pi*Z (OR=3.49, 95%CI=1.55–7.85, p-value=0.003)Likewise, AAT deficiency was associated with a higher risk for asthma exacerbations (OR=5.18, 95%CI=1.58–16.92, p-value=0.007) as well as AAT protein levels (OR= 0.72, 95%CI=0.57–0.91, p-value=0.005). The Pi*Z association with exacerbations was replicated in samples from Spaniards with two generations of Canary Islander origin (OR=3.79, p-value=0.028), and a significant association with asthma hospitalizations was found in the Finnish population (OR=1.12, p-value=0.007).
ConclusionsAAT deficiency could be a potential therapeutic target for asthma exacerbations in specific populations.
Asthma is a common chronic respiratory disease characterized by episodes of wheezing, coughing, shortness of breath, and airflow limitation, which vary over time and intensity.1 Moreover, about 75% of asthma patients also present atopy.1,2 Asthma is considered a complex disease with a multifactorial background, in which genetic factors are known to play a significant role, with heritability ranging between 35% and 95%.2 Among European countries, Spain exhibits variation in asthma prevalence, the Canary Islands having the highest prevalence (18%) compared to the rest of the Spanish regions (4% on average).3-5
While asthma symptoms may resolve spontaneously or in response to preventive medication, some patients with asthma may experience episodic flare-ups, known as exacerbations. The most widely used definition of asthma exacerbations comprises the administration of systemic corticosteroids, emergency room visits, or hospitalizations.6 These episodes not only negatively impact the patient's quality of life, long-term lung function, work productivity, and/or school attendance, but can also be life-threatening.1 Likewise, the associated socioeconomic burden extends to caregivers, the healthcare system, and the whole community.1
Among the asthma-associated genetic factors, the serpin family A member 1 gene (SERPINA1) participates in the development of several respiratory diseases, including asthma,7,8 and it is also associated with atopy in asthma patients.9SERPINA1 gene encodes alpha-1 antitrypsin (AAT), the most abundant antiprotease in the human serum,10 plays a fundamental role as a serine protease inhibitor, protecting the lower respiratory tract against neutrophil elastase, in addition to having other immunomodulatory and anti-inflammatory properties.11 When serum levels of AAT decrease below a protective threshold,12 reduced protection against elastase activity in the lung could lead to alveoli degradation.13
Different SERPINA1 gene variants are known to produce AAT deficiency (AATD), which is considered one of the most common hereditary disorders of European descent populations.14 AATD is diagnosed by measurement of AAT serum levels and characterization of AAT isoforms by protein isoelectric focusing (Pi*AAT phenotypes). Pi*M is considered the reference AAT variant, associated with normal AAT levels, while two canonical Pi*AAT variants, known as Pi*Z and Pi*S, have been classically associated with AATD.15Pi*Z variant is related to severe AATD,16 and is caused by a missense mutation (p.Glu342Lys, rs28929474) in the SERPINA1 gene, with a minor allele frequency (MAF) < 1%, reaching the highest MAF in European populations (2%). Pi*S variant is related to moderate AATD and is produced by another missense mutation of the SERPINA1 gene (p.Glu264Val, rs17580) with the highest MAF in European-descent populations (6%)17. Interestingly, the Spanish population shows the highest allele frequency for this polymorphism among the populations from the 1000 Genomes project (9.3%).18
To the best of our knowledge, the association of SERPINA1 classic genetic variants with asthma exacerbations has not been studied yet. In this sense, populations of the Canary Islands are interesting due to the elevated asthma prevalence in this region, and the unique genomic features found in the different islands.19,20 Indeed, novel defective SERPINA1 alleles have been characterized in La Palma island,21 where frequencies of Pi*S and Pi*Z alleles are higher than expected.22 In the present work, we performed the first genetic association study of SERPINA1 variants with asthma exacerbations, considering subjects with asthma from La Palma Island as a model. In addition, were attempted to replicate our findings in Canary Islanders, mainland Spanish subjects, and other European populations.
MethodsDiscovery and replication populationsPatients enrolled in Characterizing Alpha-1-Antitrypsin Deficiency in patients with pulmonary diseases (CAATDPUL)23 study were included in the discovery cohort. The CAATDPUL study was approved by the Ethics Committee of La Palma General Hospital, with the reference/date HGLaPalma_2010_7/ September 26, 2010. Patients enrolled in The Genomics and Metagenomics of Asthma Severity (GEMAS)24 cohort and in the Study of the mechanisms involved in the genesis and evolution of asthma (MEGA)25 were included in the replication phase. The GEMAS and MEGA studies were approved by the respective Clinical Research Ethics Committee of participating centers (approvals 29/17 for the Canary Islands hospitals and PI2019077 Hospital Universitario Donostia in the GEMAS study, and EOH 2014/48 from Hospital Universitario Fundación Jiménez Díaz in the MEGA study). All patients under 18 years old provided their assent (with signature for those aged 12–17 years), and one of their legal guardians signed the informed consent. All participants older than 18 years old provided their informed consent to participate in the study. A full description of each cohort can be found elsewhere.23-25
Details related to AAT quantification, genomic DNA extraction, genotyping, and quality control (e-Appendix 1) are described in the Supplementary Material. Moreover, the open-access integrative resource Open Targets Genetics (version 7),26 which integrates summary results of genetic association studies of the GWAS Catalog, FinnGen, and UK Biobank studies, was used for in silico assessment of replication in other European populations.
Statistical analysesSubjects who experienced hospitalizations, emergency room visits, and/or oral corticosteroids in the last 12 months were selected as cases.6 In contrast, controls did not experience any of these events for the same period. The definition of exacerbations was identical for the discovery and replication populations. The association of AATD, AAT levels, or SERPINA1 genetic variants with asthma exacerbations was tested through logistic regression models following an additive genetic model for the rs17580 and rs28929474 polymorphisms.
Deficient SERPINA1 genotypes (Pi*MS, Pi*SS, Pi*MZ, Pi*SZ, and Pi*ZZ) were compared with the reference genotype (Pi*MM). AATD was defined as a binary variable based on the AAT protein levels (<80 or ≥80 mg/dl).27 AAT levels were also analyzed as a continuous variable with a Box-Cox normalization to obtain a normal distribution. Age, sex, and PCs from the genotype data that summarized most of the genetic variability in allele frequencies were included in the regression models as covariates. The 29 subjects simultaneously genotyped from dried blood spots and fresh blood were considered in CAATDPUL and removed from further analysis in GEMAS. Statistical significance was declared after applying a Bonferroni correction for the two genetic variants tested (p-value ≤ 0.025). Sensitivity analyses were conducted and are explained in e-Appendix 2.
In the replication phase, association analyses were performed following the same methodology as in the discovery phase. In the GEMAS study, replication analyses were conducted stratifying the subjects by having or not two generations of Canary Islander origin. All analyses were performed using the statistical program R 3.6.3.28
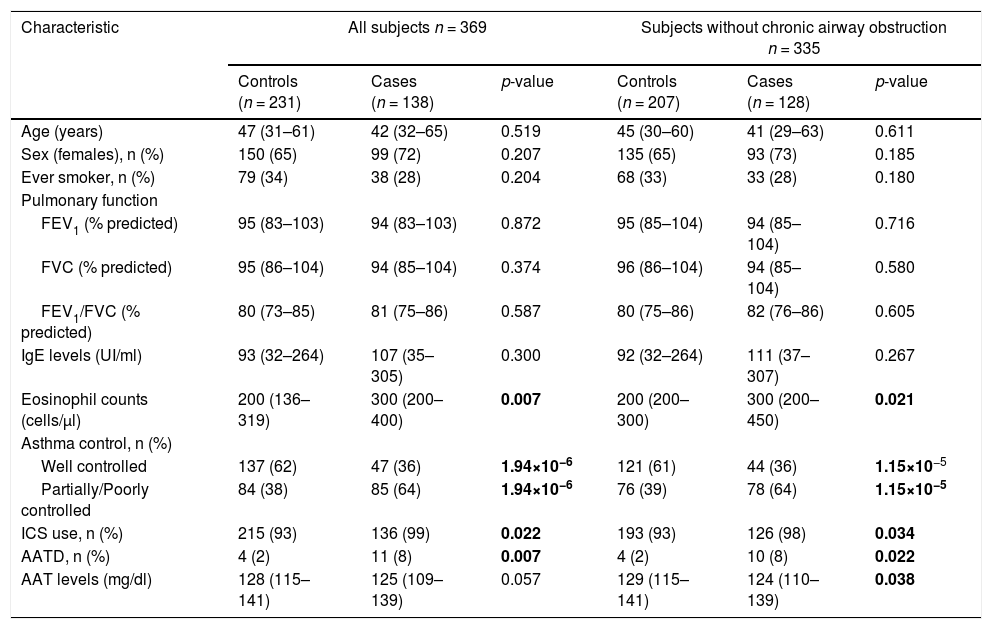
ResultsQuality control, demographic, and clinical characteristics of all study populationsAfter QC, 369 subjects from CAATDPUL, 383 from GEMAS, and 138 from MEGA populations were retained for subsequent analyses (e-Appendix 2, e-Figure 1). For subjects simultaneously genotyped from dried blood spots and fresh blood, we observed a concordance rate of 99.2%, confirming the suitability of dried blood spot samples for genome-wide genotyping without loss of accuracy and coverage (total genotyping rate of 99.7%). The main demographic and clinical characteristics of the discovery population are shown in Table 1. No significant differences were found between cases and controls for age, sex, smoking behavior, lung function measurements, and total IgE levels. Cases showed higher eosinophils levels (p-value=0.007), had a higher proportion of subjects requiring inhaled corticosteroids (ICS) (p-value=0.022), and more patients with AATD in comparison with controls (p-value=0.007). In contrast, controls showed higher AAT levels when subjects with chronic airway obstruction were excluded (p-value=0.038) and a higher proportion of controlled asthma (p-value<0.01).
Demographic and clinical variables of subjects from La Palma Island (enrolled by CAATDPUL between 2011 and 2015) included in the discovery phase.
A comparison of demographic and clinical characteristics between controls and cases was performed for all CAATDPUL subjects or excluding patients with chronic airway obstruction. The normality of continuous variables (age, pulmonary function variables, IgE levels, eosinophils counts, and AAT levels) was evaluated using the Shapiro-Wilk test and then summarized with the median and interquartile range (in brackets). Categorical variables (sex, ever smoker, asthma control, ICS use, and AATD) were summarized as counts for each group and percentages (in brackets). Predicted values of lung function measurements were estimated using the Global Lung Function Initiative (GLI) 2012 equations. Statistically significant differences between groups were evaluated either with Mann-Whitney U or Fisher's exact tests, for continuous and categorical variables, respectively. Statistically significant differences between groups (p-values<0.05) are depicted in boldface. Abbreviations: AATD: Alpha-1-antitrypsin deficiency; ACT: Asthma control test; FEV1: Forced expiratory volume in the first second; FVC: Forced vital capacity; ICS: inhaled corticosteroid; IgE: Immunoglobulin E; n: sample size.
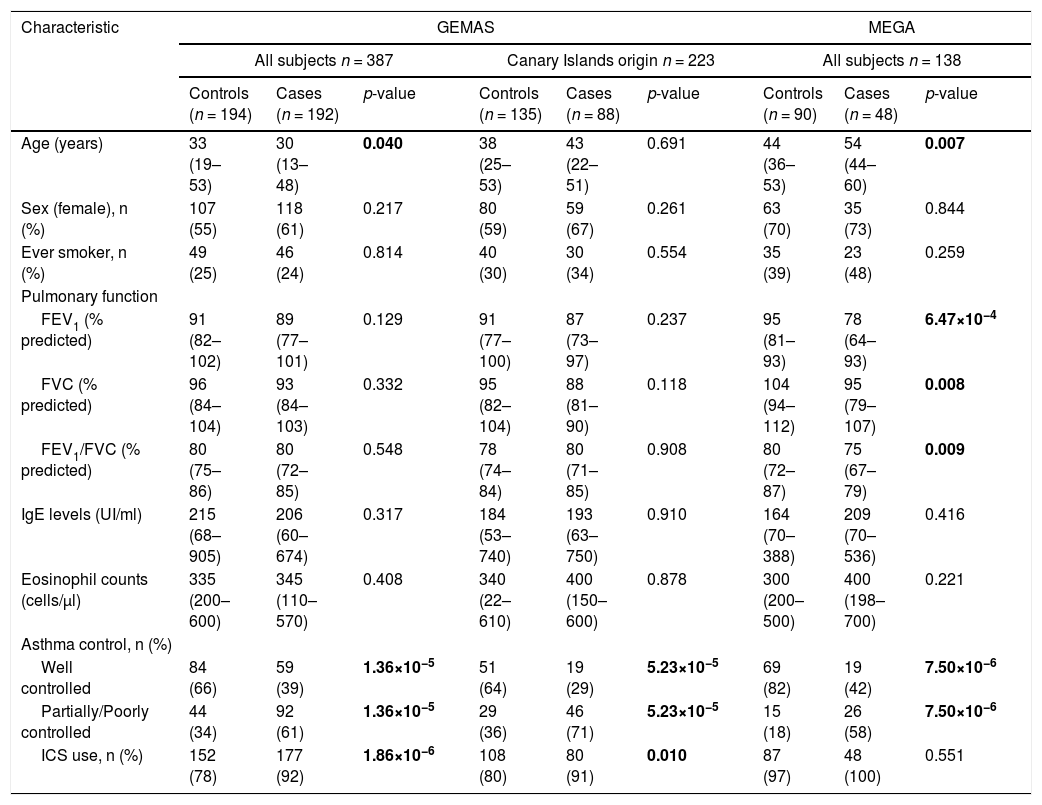
Demographic and clinical characteristics available for the two replication datasets (GEMAS and MEGA) are shown in Table 2. In the GEMAS whole study, controls were slightly older than cases (p-value=0.04), while this difference was not found in subjects with two generations of Canary Islands origin. In contrast, in the MEGA study, cases were older than controls (p-value=0.007). Similar to CAATDPUL, GEMAS cases had a higher proportion of ICS use than controls (p-value<0.001), but no difference was observed in MEGA. In addition, cases showed lower pulmonary function than controls in the MEGA population (p-value<0.01). Finally, cases presented a higher proportion of asthma partially or poorly controlled in all populations (p-value<0.001).
Demographic and clinical variables in the replication populations (GEMAS and MEGA).
Comparison of demographic and clinical characteristics between controls and cases was performed for GEMAS (including all subjects, or only patients with two generations of Canary Islander origin), and MEGA populations. The normality of continuous variables (age, pulmonary function variables, IgE levels, and eosinophils counts) was evaluated with the Shapiro-Wilk test, and then summarized with the median and interquartile range (in brackets). Categorical variables (sex, ever smoker, asthma control, and ICS use) were summarized as counts for each group and percentages (in brackets). Predicted values of lung function measurements were estimated using the Global Lung Function Initiative (GLI) 2012 equations.
Statistically significant differences between groups were evaluated either with Mann-Whitney U or Fisher's exact tests, for continuous and categorical variables, respectively. Statistically significant differences between groups (p-values<0.05) are depicted in boldface. Abbreviations: ACT: Asthma control test; FEV1: Forced expiratory volume in the first second; FVC: Forced vital capacity; ICS: inhaled corticosteroid; IgE: Immunoglobulin E; n: sample size.
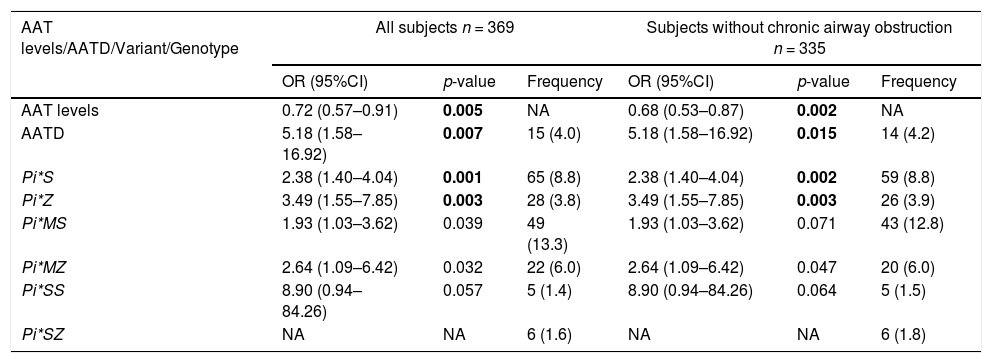
In the CAATDPUL cohort, a significant association of AAT protein levels was found, showing higher levels a protective effect against asthma exacerbation (odds ratio [OR]= 0.72, 95%CI=0.57–0.91, p-value=0.005). AATD was associated with an increased risk for asthma exacerbations (OR=5.18, 95% CI: 1.58–16.92, p-value=0.007). SERPINA1 deficient variants Pi*S (OR=2.38, 95%CI=1.40–4.04, p-value=0.001) and Pi*Z (OR=3.49, 95%CI=1.55–7.85, p-value=0.003) were associated with asthma exacerbations (Table 3). Likewise, the associations of Pi*S and Pi*Z alleles, AAT levels, and AATD with asthma exacerbations were consistent in the analyses that excluded subjects with chronic airflow obstruction. Moreover, all the associations retained their significance in models adjusted for smoking history, BMI, and ICS use (e-Table 1).
Association analysis results obtained between asthma exacerbations and SERPINA1 genetic variants, classic genotypes, and AATD, as well as their frequencies in the discovery population and adjusting by age, sex, and principal components.
AATD frequency was summarized as counts and% frequency (in brackets). The frequency of variants and genotypes were summarized as allele/genotype count and minor allele frequency/genotype percentages (in brackets). Statistically significant p-values after Bonferroni correction are depicted in boldface (p-value<0.025). Abbreviations: AATD: Alpha-1 antitrypsin deficiency; CI: Confidence interval; NA: Not available (beta coefficient or standard deviation >10); n: sample size; OR: Odds ratio.
In the assessment of alternative outcomes, associations were found (e-Table 2) for AATD with ACT level (p-value=0.017) and Pi*MS genotype with asthma severity (p-value=0.014), but not for other variables.
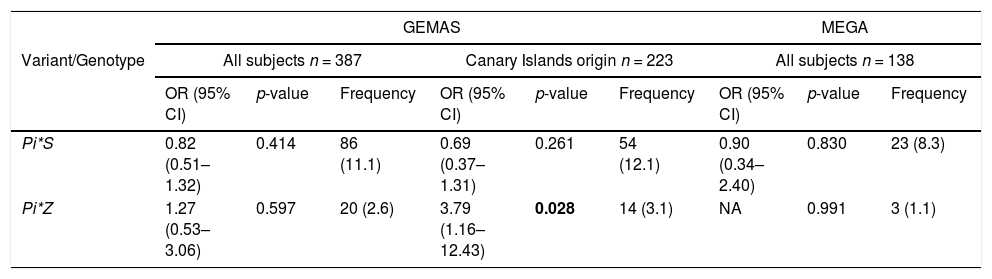
Association analysis in the replication phaseWe next examined deficient variants for association with asthma exacerbations in the replication cohorts. The association between the Pi*Z variant and asthma exacerbations was nominally replicated in a subset of GEMAS patients that had four grandparents from the Canary Islands (OR=3.79, 95%CI=1.16–12.43, p-value=0.028) (Table 4). However, the association of Pi*S variant with asthma exacerbations was not significant in this subset of patients. Moreover, none of the associations between deficient variants and asthma exacerbations were validated when all subjects from GEMAS or MEGA were considered (Table 4).
Association of SERPINA1 genetic variants and genotypes with exacerbations and their frequencies in the replication populations and adjusting by age, sex, and principal components.
The frequency of variants and genotypes were summarized as allele/genotype count and minor allele frequency/genotype percentages (in brackets). Statistically significant p-values are depicted in boldface (p-value<0.05). Abbreviations: CI: Confidence interval; NA: Not available (beta coefficient or standard deviation >10); n: sample size; OR: Odds ratio.
An in-silico assessment in other European populations via Open Targets Genetics revealed a significant association of the Pi*Z variant with asthma (OR=2.35, p-value=2.3 × 10−3) and only in the Finnish population (OR=1.16, p-value=1.1 × 10−4).26 In addition, a significant association of Pi*Z with asthma hospitalizations (OR=1.12, p-value=6.5 × 10−3) in Finnish population26 was found but not in other European populations.
DiscussionTo the best of our knowledge, this is the first study examining the association of AATD and SERPINA1 variants with asthma exacerbations. A higher level of AAT was associated with a protective effect for asthma exacerbations among Canary Islanders from La Palma. In addition, AATD, considered when serum protein level was below 80 mg/dl, was associated with a higher risk for asthma exacerbations. In the same population, the rs17580 (Pi*S) and rs28929474 (Pi*Z) SERPINA1 variants were associated with a higher risk of asthma exacerbations. The rs28929474 signal was replicated in an independent sample of the Canary Islands and Finnish populations, but not in other Spanish or European populations.
Several genes have been associated with predisposition to this flare-up episode,29-36 but in-depth studies, carried out in additional populations, are necessary to identify the whole genetic component involved in asthma exacerbations. Nevertheless, AATD is considered an underdiagnosed disease, as only about 10% of patients are estimated to be properly diagnosed.37 Therefore, the scarce AATD diagnosis bias could underestimate its association with asthma. Since AAT is the main inhibitor of neutrophilic serine proteinase and acts as an important anti-inflammatory protein with pronounced immunomodulatory activity, the link between AATD and asthma might be represented by the elastase/antielastase imbalance caused by AATD. Alternatively, the proinflammatory effect caused by AAT serum levels reduction could also underlie the association found.7 Interestingly, a previous study observed that protein levels of AAT are elevated during asthma exacerbations,38 which agrees with our findings.
Despite the important role of SERPINA1 variants in COPD,27 the associations found were not driven by this condition, as they remained significant when all asthmatic patients with pulmonary obstruction were excluded from the analysis. Moreover, the association of AATD with asthma severity was also confirmed by analyzing asthma control values as an alternative outcome. Given that IgE and eosinophil levels are related to allergic asthma,39 and the high prevalence of atopy in the Canary Islands,5 we evaluated the association of SERPINA1 variants and genotypes with these phenotypes. However, no associations were found in this case, suggesting the SERPINA1 variants are not directly involved in the immunomodulation of the allergic response in these individuals. In previous studies, the Pi*Z variant has been associated with smoking behavior17, and the Pi*S variant has been mainly associated with the risk of developing COPD in smokers.17 However, our associations of Pi*S and Pi*Z alleles, AAT levels, and AATD with asthma exacerbations were not explained by smoking history, body mass index, or ICS use.
The associations found in the discovery population motivated us to assess the validation in Spanish subjects considering the overall population and a subset of individuals with two generations of Canary Islands origin. The latter comparison allowed us to evaluate the possible existence of a population-specific effect. It is known that populations from the Canary Islands present a recent admixture involving North African, European, and sub-Saharan African ancestry with variable ancestry, depending on the island.40 Although no relationship has been found between genetic ancestry and asthma when individuals from different islands of the Canary Archipelago were analyzed,41 studies based on island-specific populations are yet to be addressed. In this sense, evidence of validation was found for the association of rs28929474 (Pi*Z) with asthma exacerbations in Canary Islanders, but not in other Spanish populations.
The rs28929474 (Pi*Z) variant was also associated with asthma hospitalizations in the Finnish population, suggesting the possibility that a founder effect in certain isolated populations could cause an increase in SERPINA1 deficient alleles associated with the risk of asthma exacerbations. In this sense, the lack of replication in the independent populations from mainland Spain may be due to differences in allele frequencies among the discovery and replication populations. In previous studies,38,42 a high frequency of both polymorphisms was found among Spanish individuals, where the Pi*S and Pi*Z variants have a frequency of 10 and 2%, respectively.43 Interestingly, the frequency of the Pi*Z variant in our study was higher in La Palma (3.8%), followed by Canary Islanders from GEMAS (3.1%), and mainland Spaniards from MEGA (1.1%). In the case of the Finnish population, the frequency of Pi*Z was only two times lower than in La Palma (1.9%), but the frequency of Pi*S was about 10 times lower (0.8%), which could explain the lack of replication of the Pi*S variant in this population.
To the best of our knowledge, the present work represents the first evidence of the association of AATD and SERPINA1 classic variants with asthma exacerbations. Moreover, we have demonstrated the robustness of the results by including strict quality control procedures and considering multiple demographic and clinical confounders in the association analyses. Additionally, we have analyzed different populations, not only genotyped in our study but also from different public databases queries.
Nonetheless, some limitations must be considered. First, the Pi*S association with asthma exacerbations was not validated in all populations analyzed in the replication stage. Although the association found could be population specific, the lack of replication could also be due to the reduced statistical power derived from the smaller sample size in the non-Canary Islander replication set of samples. Second, different endotypes of asthma were not assessed and could underlie the different results obtained in the cohorts analyzed. Third, AAT serum levels were not available in GEMAS and MEGA. Therefore, it was not possible to confirm the association between AATD and asthma exacerbations in the replication phase. Fourth, other possible confounding factors were not evaluated, for example, atopy, rhinitis, or other individual or environmental conditions, since data was not available for all cohorts, and these uncontrolled conditions could mask the associations in the replication populations. Fifth, the recruitment of patients is heterogeneous between studies since CAATDPUL enrolled patients from the Pneumonology unit only, but GEMAS and MEGA enrolled patients from Pneumonology and Allergology units. Despite correcting the regression models by the recruitment center, this feature could be another reason for the lack of replication. Additionally, differences in the environment could also affect the associations through gene-environment interactions.
Sixth, rare variants can also cause AATD, and such type of variation was not assessed. Therefore, future sequencing efforts analyzing the whole spectrum of genetic variation in the SERPINA1 gene could reveal additional associations with asthma exacerbations.
Personalized medicine has shown the existence of drugs with application to specific populations.44 In this sense, it could be helpful to measure AAT serum levels and assess the SERPINA1 genetic variants in asthmatic patients routinely in clinical practice to detect carriers of SERPINA1 deficient variants, during which implementing AAT replacement therapy could be a potential treatment to reduce asthma exacerbations. This approach would especially be relevant to patients who do not respond to other asthma treatments, in order to decrease disease morbidity and mortality.
ConclusionsWe describe for the first time the association of the SERPINA1 gene Pi*S (rs17580) and Pi*Z (rs28929474) variants, AAT levels, and AATD with asthma exacerbations in patients from La Palma (Canary Islands, Spain). In addition, the association of the Pi*Z allele, the most important SERPINA1 variant in clinical practice, was validated in another set of Canary Islanders and individuals from the Finnish population. This suggests AATD is a potential therapeutical target for treating asthma exacerbations in specific populations.
Author contributionsM.P.Y. and M.G.C. are the guarantors of the paper, taking responsibility for the integrity of the work, from inception to the published article. E.M.G. was responsible for formal analysis and writing the original draft. J.M.H.P., M.P.Y., and M.G.C. contributed to the conceptualization, and M.P.Y. and M.G.C. also contributed to methodology and supervision. Resources were carried out by J.M.H.P., J.A.P.P., R.G.P., E.M.L., I.S.M., O.S., P.C., P.P.G., J.K.M., V.d.P., F.L.D., J.V., and M.P.Y. All the authors support with the investigation, writing-review, and editing. Funding acquisition was carried out by J.M.H.P. and M.P.Y.
Financial/nonfinancial disclosuresThis study was funded by the Spanish Ministry of Science and Innovation MCIN/AEI/10.13039/501100011033 (PID2020–116274RB-I00) and by Sociedad Española de Neumología y Cirugía Torácica (SEPAR http://www.separ.es), grant number 1264–2022. Authors declare they have no competing interests or other interests that might be perceived to influence the interpretation of the manuscript. No supporting institution may gain or lose financially through this publication. Genotyping of samples from GEMAS and MEGA studies was funded by the Spanish Ministry of Science and Innovation (SAF2017–87417R) at the Spanish National Cancer Research centre, in the Human Genotyping lab, a member of CeGen, PRB3, and was supported by grant PT17/0019, of the PE I + D + i 2013–2016, funded by Instituto de Salud Carlos III (ISCIII) and European Regional Development Fund. Genotyping of GEMAS was also partially funded by Fundación Canaria Instituto de Investigación Sanitaria de Canarias (PIFIISC19/17). E.M.G. was supported by a fellowship (TESIS2022010045) awarded by the Board of Economy, Industry, Trade, and Knowledge of the Canary Islands Government, with a European Social Fund co-financing rate managed by the Canary Islands Agency for Research, Innovation, Society, and Information (ACIISI). J.P.G. was funded by an FPU fellowship (FPU19/02,175) granted by the Spanish Ministry of Universities. E.H.L. was supported by a fellowship awarded by MCIN/AEI/10.13039/501100011033 and by “ESF Investing in your future” (PRE2018–083,837). V.P. has received grant support from MSD. V.d.P. was supported by CIBER de Enfermedades Respiratorias, ISCIII, Spain. M.P.Y. was funded by the Ramón y Cajal Program (RYC-2015–17,205) by MCIN/AEI/10.13039/501100011033 and by the European Social Fund “ESF Investing in your future”. M.P.Y. and J.V. were supported by CIBER de Enfermedades Respiratorias, ISCIII, Spain (CB/06/06/1088).
Role of sponsorsThe sponsor had no role in the design of the study, the collection, and analysis of the data, or the preparation of the manuscript.
Other contributionsThe authors thank the patients, families, recruiters, healthcare providers, and community clinics for their participation in the studies analyzed in this manuscript. The genotyping service was carried out at CEGEN-PRB3-ISCIII; it is supported by grant PT17/0019, of the PE I + D + i 2013–2016, funded by ISCIII and ERDF.
FundingThis study was funded by the Spanish Ministry of Science and Innovation MCIN/AEI/10.13039/501100011033 (PID2020–116274RB-I00) and by Sociedad Española de Neumología y Cirugía Torácica (SEPAR http://www.separ.es), grant number 1264–2022.