Hypersensitivity pneumonitis (HP) is an interstitial lung disease with diverse clinical features that can present a fibrotic phenotype similar to idiopathic pulmonary fibrosis (IPF) in genetically predisposed individuals. While several single nucleotide polymorphisms (SNPs) have been associated with IPF, the genetic factors contributing to fibrotic HP (fHP) remain poorly understood. This study investigated the association of MUC5B and TOLLIP variants with susceptibility, clinical presentation and survival in Portuguese patients with fHP.
Material and MethodsA case-control study was undertaken with 97 fHP patients and 112 controls. Six SNPs residing in the MUC5B and TOLLIP genes and their haplotypes were analyzed. Associations with risk, survival, and clinical, radiographic, and pathological features of fHP were probed through comparisons among patients and controls.
ResultsMUC5B rs35705950 and three neighboring TOLLIP variants (rs3750920, rs111521887, and rs5743894) were associated with increased susceptibility to fHP. Minor allele frequencies were greater among fHP patients than in controls (40.7% vs 12.1%, P<0.0001; 52.6% vs 40.2%, P = 0.011; 22.7% vs 13.4%, P = 0.013; and 23.2% vs 12.9%, P = 0.006, respectively). Haplotypes formed by these variants were also linked to fHP susceptibility. Moreover, carriers of a specific haplotype (G-T-G-C) had a significant decrease in survival (adjusted hazard ratio 6.92, 95% CI 1.73–27.64, P = 0.006). Additional associations were found between TOLLIP rs111521887 and rs5743894 variants and decreased lung function at baseline, and the MUC5B SNP and radiographic features, further highlighting the influence of genetic factors in fHP.
ConclusionThese findings suggest that TOLLIP and MUC5B variants and haplotypes may serve as valuable tools for risk assessment and prognosis in fibrotic hypersensitivity pneumonitis, potentially contributing to its patient stratification, and offer insights into the genetic factors influencing the clinical course of the condition.
Hypersensitivity pneumonitis (HP) is an interstitial lung disease (ILD), with a heterogeneous clinical presentation and evolution, recently classified as comprising two forms – fibrotic and non-fibrotic.1-3 HP is triggered by exposure to a variety of inciting antigens that results in inflammation, causes persistent lung injury and, in genetically predisposed individuals, leads to fibrotic disease, similar to idiopathic pulmonary fibrosis (IPF).4,5 The differential diagnosis of these two fibrosing entities is challenging but imperative given the therapeutic and prognostic implications.1,4,6,7 In fibrotic HP (fHP), antigen avoidance, if recognized, is the mainstay of treatment, associated with immunosuppression, when necessary.8,9 Antifibrotics, the standard treatment for IPF, are only prescribed later in cases of HP with progressive pulmonary fibrosis.10-12
Several single nucleotide polymorphisms (SNPs) have been associated with IPF susceptibility and survival, such as the mucin 5B (MUC5B) rs35705950 promoter polymorphism and several toll-interacting protein (TOLLIP) SNPs.13-17 Whether independently associated or integrating haplotypes potentially predicting altered survival,18 these variants highlight the contribution of genetic determinants to the development and evolution of the fibrosing phenotype.
In fHP, in contrast, very little is known about predisposing genetic factors.19 To date, no genome-wide association studies have been performed although a few studies probing candidates chosen on the basis of association with other ILDs have identified associations with disease susceptibility and outcomes in fHP.20-22 Among those, the MUC5B promoter variant and telomere length were implicated as susceptibility and survival determinants in patients with chronic HP,20,23,24 as extensively reported for IPF.25 No study has yet focused on the involvement of the various IPF-related TOLLIP SNPs in the context of fHP.
The objective of this study was to determine whether the MUC5B promoter and neighboring TOLLIP polymorphisms, and respective haplotypes, are associated with susceptibility, clinical presentation and survival in patients with fHP.
Materials and methodsParticipants and study designA case-control study was conducted with 97 non-familial fHP patients followed from 2013 to 2022 in a tertiary Portuguese hospital in Porto. This group included cases enrolled in the FIBRALUNG (Fibrosing ILD Biomarkers That Rule Acceleration) project, the first national registry and biobank of ILDs.26 A control group with 112 subjects was selected from EPIPorto,27 a population-based cohort of healthy adults (n = 74), and NETDiamond,28,29 a prospective cohort for the study of therapeutic targets in heart failure, selecting those without respiratory disease (n = 38).
Diagnosis of fHP was established according to international guidelines.1,2,6 Demographic and clinical information, including lung function, bronchoalveolar lavage (BAL) as well as radiographic and histopathological data, were collected on enrolment.
The study protocol was reviewed and approved by the Ethics Committee of the Institution and performed in accordance with the Helsinki Declaration. Written informed consent was obtained from all participants.
GenotypingIndividuals were genotyped for six SNPs: TOLLIP rs3750920, rs111521887, rs5743894, rs5743890, rs5743854, and MUC5B rs35705950, as described,18 and by RFLP and real-time PCR (details in Supplementary Methods M1, Table S1).
RadiologyHigh-resolution computed tomography (HRCT) scans were independently reviewed by a radiologist with experience in ILD CT scan interpretation, blinded to clinical diagnosis. Detailed information on image analysis and classification scores can be found in Supplementary Methods M2.
HistopathologyLung biopsies were scored and reviewed by an expert lung pathologist in ILDs (surgical lung biopsy or transbronchial lung cryobiopsy) blinded to the original diagnosis, using a guided pathology data collection form. Detailed information on histopathological analyses and classification scores can be found in Supplementary Methods M3.
Statistical analysisStatistical analyses were performed in R (v4.2.0). Haplotype frequencies were estimated using Haploview v4.2. Pairwise linkage disequilibrium (LD) between SNPs was calculated using SNPAlyze v8.1.1.0 (Dynacom Co., Japan).
Continuous data are presented as means ± standard deviation or medians and interquartile range. Categorical data are presented as frequencies and percentages. Chi-squared tests were used to compare allele and genotype frequencies between patients and controls.
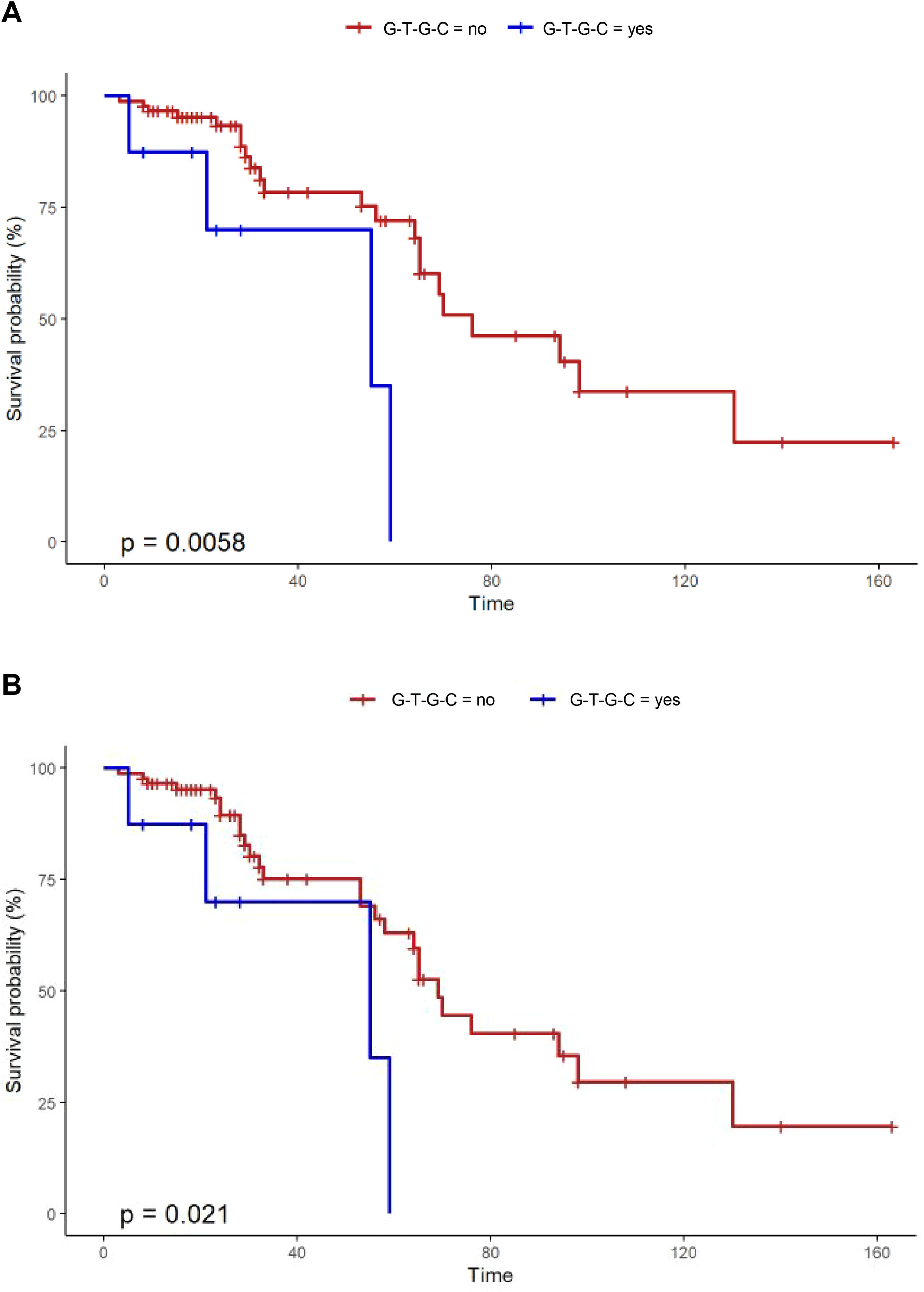
Overall survival was analyzed with time from diagnosis to death (in months) censured by end of follow-up or lung transplantation. Transplant-free survival considered lung transplantation equivalent to death. Kaplan-Meier plots and log-rank tests were used for survival and transplant-free survival.
Univariate and multivariate Cox proportional hazards regressions were employed to identify variables that predict survival status, with results being expressed as hazard ratio (HR) and respective 95% confidence interval (CI).
Two-sided P-values lower than 0.05 were considered statistically significant.
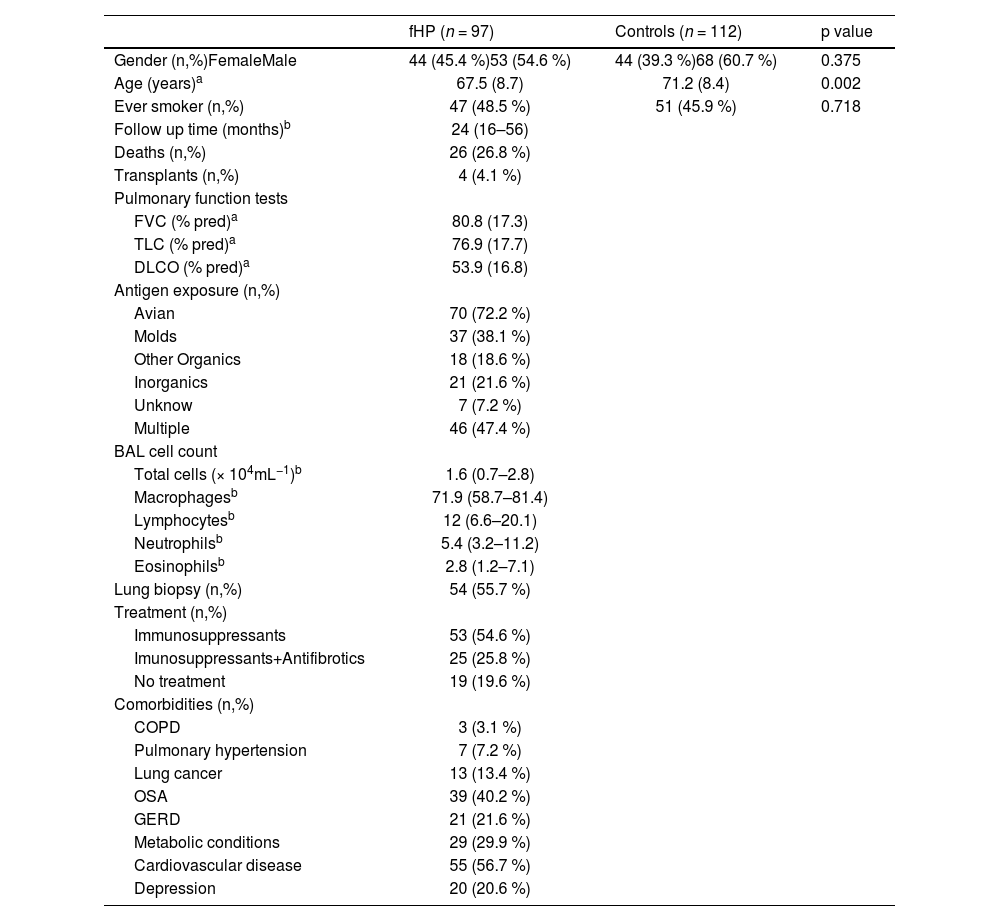
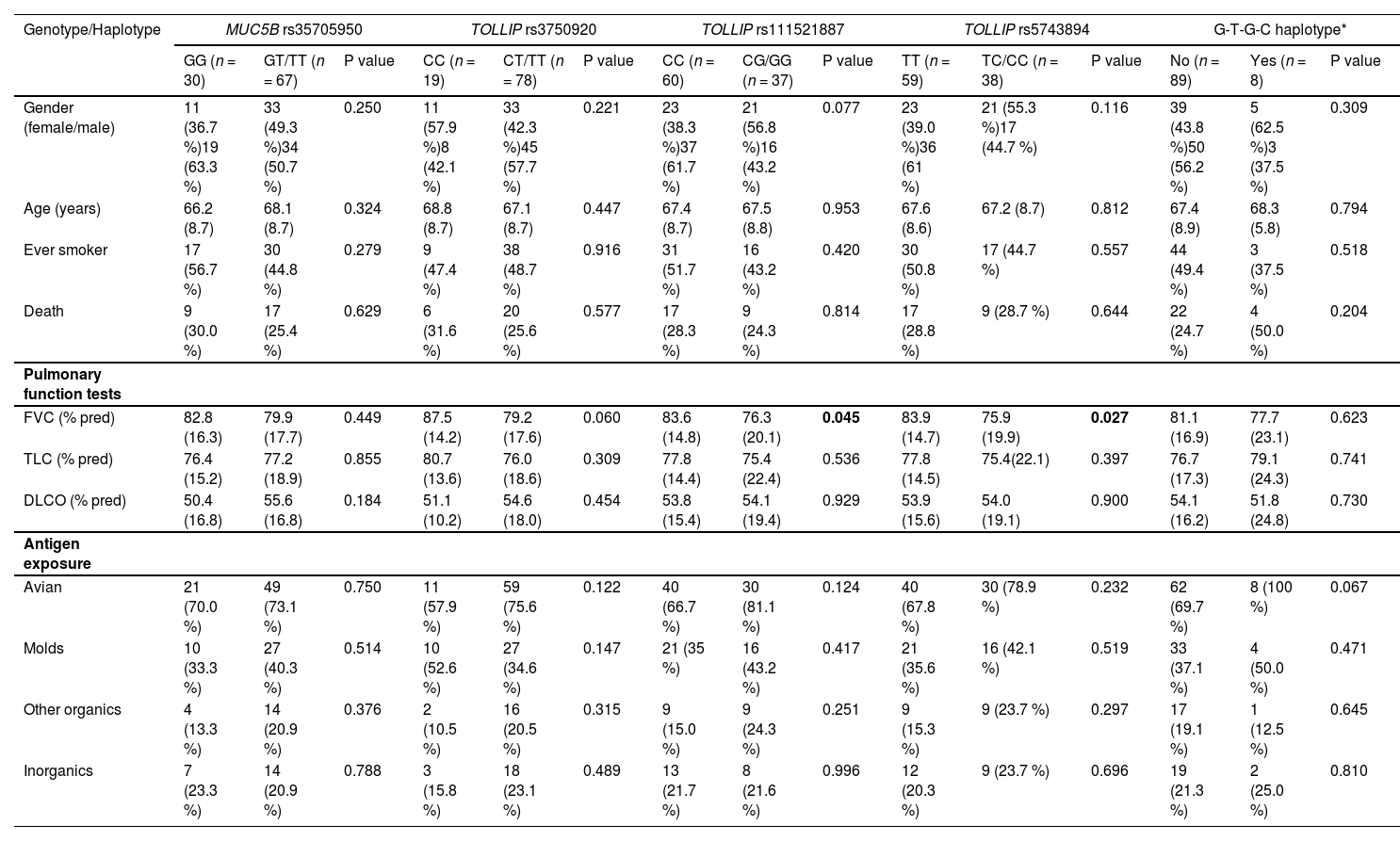
ResultsDemographic and clinical features of the study subjectsThe baseline characteristics of participants are shown in Table 1. There were no significant differences between the groups regarding gender and smoking history, although fHP patients were younger than control subjects (mean 67.5 vs 71.2 years, P = 0.002). Antigen exposure was identified in 93% of patients, with avian and molds being the most frequently reported.
Clinical and demographic characteristics of fHP patients and controls.
Values are presented as amean (±standard deviation), bmedian (Q1-Q3) and number (%).
fHP, fibrotic hypersensitivity pneumonitis; COPD, chronic obstructive lung disease; OSA, obstructive sleep apnea; GERD, gastro-esophageal reflux disease;% pred, percentage of predicted normal value; FVC, forced vital capacity; TLC, total lung capacity; DLCO, diffusing capacity of the lung for carbon monoxide.
All patients had radiographic signs of fibrosis. Histological confirmation of fHP diagnosis was necessary in 55.7% of patients. Most patients were initially treated with immunosuppressants with the addition of antifibrotics in cases of progressive pulmonary fibrosis.
Overall median follow-up duration, defined as time from diagnosis to death/transplant or end of study, was 24 months. Twenty-six patients (26.8%) died during follow-up.
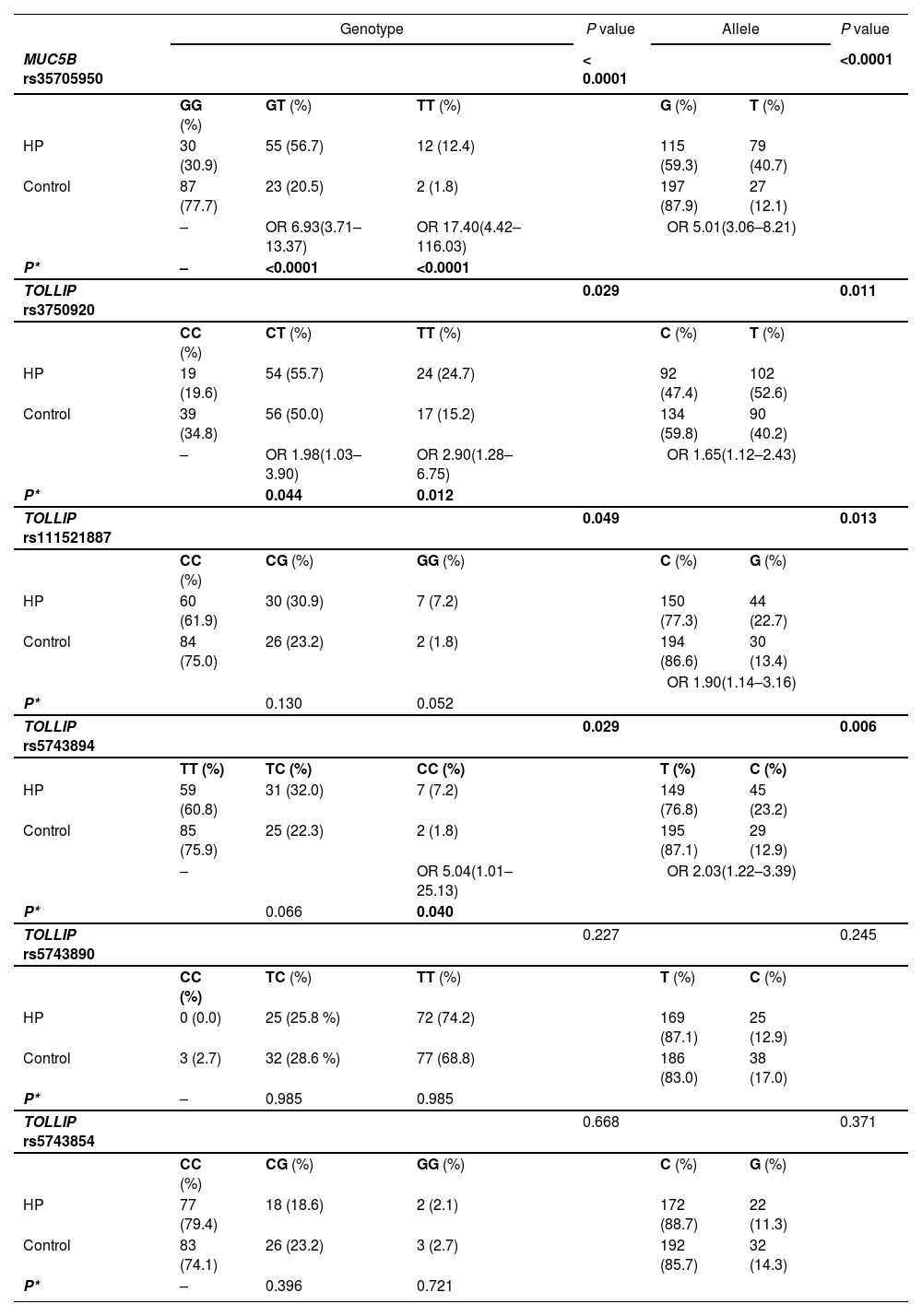
MUC5B and TOLLIP associations with fHP susceptibility and survivalGenotype frequencies of the probed SNPs are distributed as shown in Table 2, all conforming to the Hardy-Weinberg equilibrium (Supplementary Table S2).
Genotype and allele distributions of MUC5B and TOLLIP SNPs in fHP patients and controls.
OR, Odds Ratio (95 % Confidence Interval).
P*, Logistic regression P value of each minor allele genotype vs. reference genotype (ancestral allele).
Alleles are reported in the Forward strand orientation, except rs5743854 in conformity with the dbSNP database minor allele designation.
The frequency of the MUC5B rs35705950 minor allele T was significantly increased in fHP patients (40.7% vs 12.1%, OR=5.01, 95% CI 3.06–8.21, P < 0.0001). Accordingly, the genotypes of T allele carriers were significantly more frequent in the fHP group (69.1% vs 22.3 %, OR=7.77, 95 % CI 4.18-14.43, P < 0.0001), indicating this variant allele also confers susceptibility to fHP.
TOLLIP rs3750920, rs111521887 and rs5743894 SNPs (henceforth also referred to as T920, T887 and T894) were significantly associated with fHP. The minor allele frequencies (MAF) were significantly increased in fHP patients relative to those of controls (52.6% vs 40.2%, P = 0.011; 22.7% vs 13.4%, P = 0.013; and 23.2% vs 12.9%, P = 0.006; respectively). The overall genotypic distributions of these SNPs were also associated with fHP (P = 0.029, P = 0.049, P = 0.029), with homozygotes bearing the minor alleles at higher risk of developing the disease (significantly so for T920 TT OR=2.90, P = 0.012; and T894 CC OR=5.04, P = 0.040; and borderline significant for T887 GG, P = 0.052). Allele and genotype distributions of TOLLIP rs5743890 and rs5743854 SNPs did not differ significantly between patients and controls.
Kaplan-Meier survival analysis of MUC5B and TOLLIP SNPs did not show any association between individual genotypes and survival in fHP patients (Supplementary Fig. S1).
MUC5B and TOLLIP haplotype associations with fHP susceptibility and survivalHaplotypes defined by the MUC5B and three neighboring TOLLIP SNPs that had shown an association with susceptibility to fHP (MUC5B-T920-T887-T894 haplotype block) were reconstructed (Supplementary Table S3). Six major haplotypes were defined, of which five showed an association with susceptibility to the disease. Haplotypes T-T-C-T, T-T-G-C, and T-C-C-T were significantly more frequent, and haplotypes G-C-C-T and G-T-C-T were significantly less frequent in patients than in controls.
Pairwise LD measures indicated moderate to strong LD among the TOLLIP SNPs and moderate LD between these and the MUC5B SNP in the fHP group, while in controls linkage between tightly linked T887 and T894 and the T920 SNP was only moderate, and with the MUC5B SNP was considerably low (Supplementary Table S4 and Fig. S2).
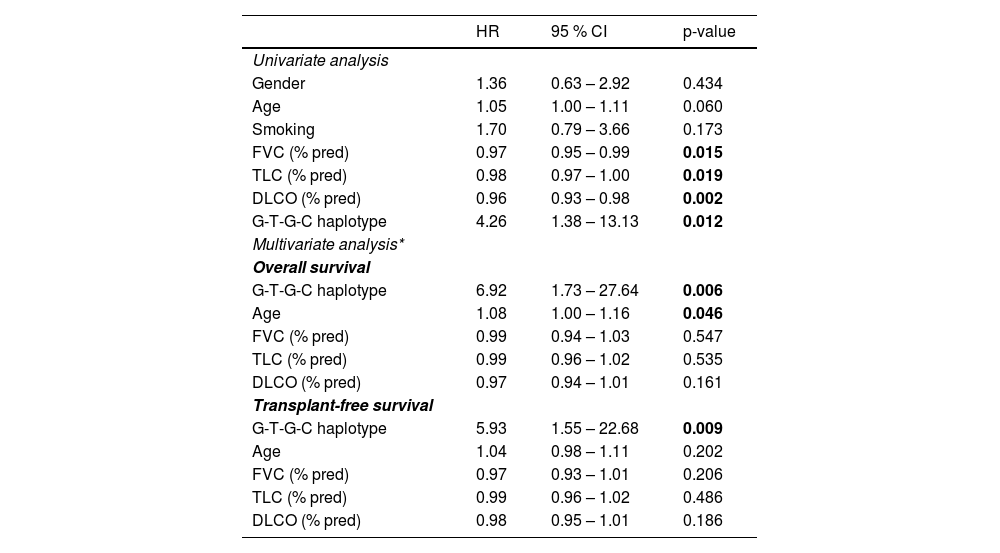
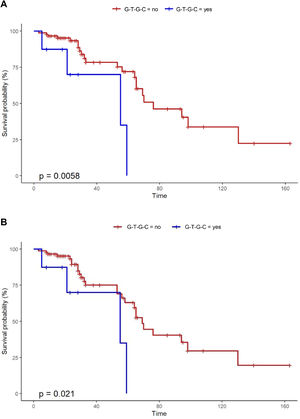
Notably, Kaplan-Meier survival analysis of the haplotypes revealed a statistically significant association of haplotype G-T-G-C with reduced survival (censoring for lung transplant) (log rank test P-value = 0.006) and reduced transplant-free survival (log rank test P-value = 0.021) in fHP (Fig. 1). Multivariate Cox regression analysis showed that the presence of the G-T-G-C haplotype was independently associated with shorter survival after adjustment for age and lung function parameters (overall survival, HR 6.92, P = 0.006; transplant-free survival, HR 5.93, P = 0.009) (Table 3). Despite being a relatively low frequency haplotype, the difference in median survival time between non-carriers and carriers was 21 months.
Univariate and multivariate Cox proportional hazard regressions predicting patients’ death.
| HR | 95 % CI | p-value | |
|---|---|---|---|
| Univariate analysis | |||
| Gender | 1.36 | 0.63 – 2.92 | 0.434 |
| Age | 1.05 | 1.00 – 1.11 | 0.060 |
| Smoking | 1.70 | 0.79 – 3.66 | 0.173 |
| FVC (% pred) | 0.97 | 0.95 – 0.99 | 0.015 |
| TLC (% pred) | 0.98 | 0.97 – 1.00 | 0.019 |
| DLCO (% pred) | 0.96 | 0.93 – 0.98 | 0.002 |
| G-T-G-C haplotype | 4.26 | 1.38 – 13.13 | 0.012 |
| Multivariate analysis* | |||
| Overall survival | |||
| G-T-G-C haplotype | 6.92 | 1.73 – 27.64 | 0.006 |
| Age | 1.08 | 1.00 – 1.16 | 0.046 |
| FVC (% pred) | 0.99 | 0.94 – 1.03 | 0.547 |
| TLC (% pred) | 0.99 | 0.96 – 1.02 | 0.535 |
| DLCO (% pred) | 0.97 | 0.94 – 1.01 | 0.161 |
| Transplant-free survival | |||
| G-T-G-C haplotype | 5.93 | 1.55 – 22.68 | 0.009 |
| Age | 1.04 | 0.98 – 1.11 | 0.202 |
| FVC (% pred) | 0.97 | 0.93 – 1.01 | 0.206 |
| TLC (% pred) | 0.99 | 0.96 – 1.02 | 0.486 |
| DLCO (% pred) | 0.98 | 0.95 – 1.01 | 0.186 |
HR, Hazard Ratio; 95 % CI, Confidence Interval.
% pred, percentage of predicted normal value.
FVC, Forced Vital Capacity; TLC, Total Lung Capacity; DLCO, Diffusing Capacity of the Lung for Carbon Monoxide.
Univariate analyses did not reveal associations with gender, age, and smoking. Conversely, better pulmonary function (FVC, TLC and DLCO) associated significantly with longer survival (HR 0.97, P = 0.015; HR 0.98 P = 0.019 and HR 0.96, P = 0.002; respectively) (Table 3).
MUC5B and TOLLIP associations with pulmonary function and clinical featuresThe presence of at least one minor allele of TOLLIP SNPs T887 and T894 was associated with lower baseline FVC% pred (genotypes GG/CG vs. CC, 76.3% vs 83.6%, P = 0.045; and genotypes CC/CT versus TT, 75.9% vs 83.9%, P = 0.027; respectively), whereas for T920 it was of borderline statistical significance (79.2% vs 87.5%, P = 0.060) (Table 4). MUC5B rs35705950 and the TOLLIP T890 and T854 polymorphisms showed no such association (Table 4 and Supplementary Table S5).
MUC5B and TOLLIP associations with demographic and clinical features.
| Genotype/Haplotype | MUC5B rs35705950 | TOLLIP rs3750920 | TOLLIP rs111521887 | TOLLIP rs5743894 | G-T-G-C haplotype* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG (n = 30) | GT/TT (n = 67) | P value | CC (n = 19) | CT/TT (n = 78) | P value | CC (n = 60) | CG/GG (n = 37) | P value | TT (n = 59) | TC/CC (n = 38) | P value | No (n = 89) | Yes (n = 8) | P value | |
| Gender (female/male) | 11 (36.7 %)19 (63.3 %) | 33 (49.3 %)34 (50.7 %) | 0.250 | 11 (57.9 %)8 (42.1 %) | 33 (42.3 %)45 (57.7 %) | 0.221 | 23 (38.3 %)37 (61.7 %) | 21 (56.8 %)16 (43.2 %) | 0.077 | 23 (39.0 %)36 (61 %) | 21 (55.3 %)17 (44.7 %) | 0.116 | 39 (43.8 %)50 (56.2 %) | 5 (62.5 %)3 (37.5 %) | 0.309 |
| Age (years) | 66.2 (8.7) | 68.1 (8.7) | 0.324 | 68.8 (8.7) | 67.1 (8.7) | 0.447 | 67.4 (8.7) | 67.5 (8.8) | 0.953 | 67.6 (8.6) | 67.2 (8.7) | 0.812 | 67.4 (8.9) | 68.3 (5.8) | 0.794 |
| Ever smoker | 17 (56.7 %) | 30 (44.8 %) | 0.279 | 9 (47.4 %) | 38 (48.7 %) | 0.916 | 31 (51.7 %) | 16 (43.2 %) | 0.420 | 30 (50.8 %) | 17 (44.7 %) | 0.557 | 44 (49.4 %) | 3 (37.5 %) | 0.518 |
| Death | 9 (30.0 %) | 17 (25.4 %) | 0.629 | 6 (31.6 %) | 20 (25.6 %) | 0.577 | 17 (28.3 %) | 9 (24.3 %) | 0.814 | 17 (28.8 %) | 9 (28.7 %) | 0.644 | 22 (24.7 %) | 4 (50.0 %) | 0.204 |
| Pulmonary function tests | |||||||||||||||
| FVC (% pred) | 82.8 (16.3) | 79.9 (17.7) | 0.449 | 87.5 (14.2) | 79.2 (17.6) | 0.060 | 83.6 (14.8) | 76.3 (20.1) | 0.045 | 83.9 (14.7) | 75.9 (19.9) | 0.027 | 81.1 (16.9) | 77.7 (23.1) | 0.623 |
| TLC (% pred) | 76.4 (15.2) | 77.2 (18.9) | 0.855 | 80.7 (13.6) | 76.0 (18.6) | 0.309 | 77.8 (14.4) | 75.4 (22.4) | 0.536 | 77.8 (14.5) | 75.4(22.1) | 0.397 | 76.7 (17.3) | 79.1 (24.3) | 0.741 |
| DLCO (% pred) | 50.4 (16.8) | 55.6 (16.8) | 0.184 | 51.1 (10.2) | 54.6 (18.0) | 0.454 | 53.8 (15.4) | 54.1 (19.4) | 0.929 | 53.9 (15.6) | 54.0 (19.1) | 0.900 | 54.1 (16.2) | 51.8 (24.8) | 0.730 |
| Antigen exposure | |||||||||||||||
| Avian | 21 (70.0 %) | 49 (73.1 %) | 0.750 | 11 (57.9 %) | 59 (75.6 %) | 0.122 | 40 (66.7 %) | 30 (81.1 %) | 0.124 | 40 (67.8 %) | 30 (78.9 %) | 0.232 | 62 (69.7 %) | 8 (100 %) | 0.067 |
| Molds | 10 (33.3 %) | 27 (40.3 %) | 0.514 | 10 (52.6 %) | 27 (34.6 %) | 0.147 | 21 (35 %) | 16 (43.2 %) | 0.417 | 21 (35.6 %) | 16 (42.1 %) | 0.519 | 33 (37.1 %) | 4 (50.0 %) | 0.471 |
| Other organics | 4 (13.3 %) | 14 (20.9 %) | 0.376 | 2 (10.5 %) | 16 (20.5 %) | 0.315 | 9 (15.0 %) | 9 (24.3 %) | 0.251 | 9 (15.3 %) | 9 (23.7 %) | 0.297 | 17 (19.1 %) | 1 (12.5 %) | 0.645 |
| Inorganics | 7 (23.3 %) | 14 (20.9 %) | 0.788 | 3 (15.8 %) | 18 (23.1 %) | 0.489 | 13 (21.7 %) | 8 (21.6 %) | 0.996 | 12 (20.3 %) | 9 (23.7 %) | 0.696 | 19 (21.3 %) | 2 (25.0 %) | 0.810 |
Values are presented as mean (SD) and counts as n (%).
Exposure to birds as an inciting antigen was tendentially more frequent in carriers of the G-T-G-C haplotype (P = 0.067). Indeed, all carriers of the haplotype were exposed to birds, whereas 30.3% of the non-carriers were not (Table 4).
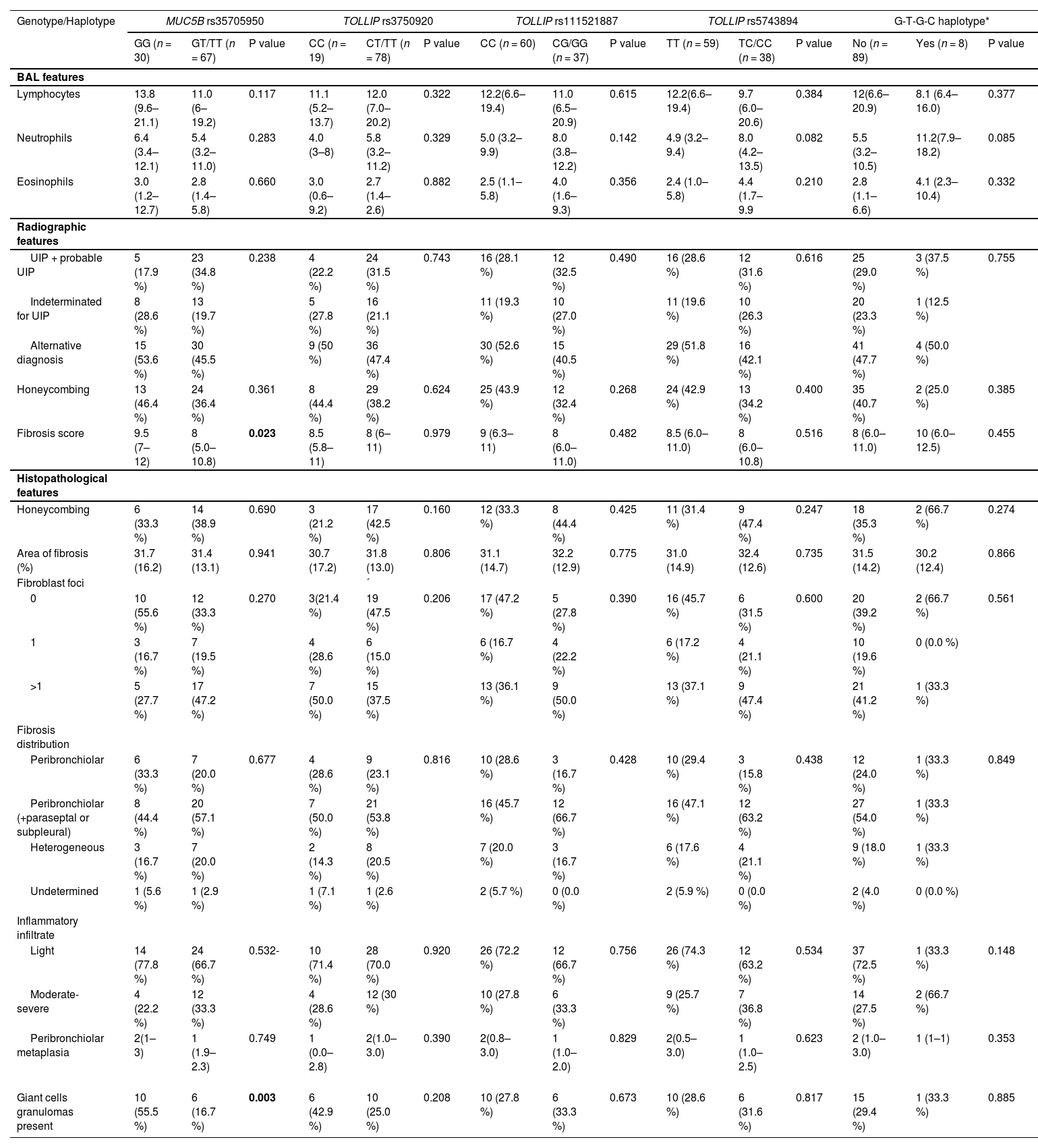
Radiographic analyses showed that MUC5B rs35705950 minor alleles were associated with a lower total score of fibrosis (P = 0.023), but not to honeycombing or a pattern indicative of definite or probable UIP. No associations between radiographic features and genotype distributions were seen for the TOLLIP SNPs (Table 5).
MUC5B and TOLLIP associations with BAL, radiographic and histopathological features.
| Genotype/Haplotype | MUC5B rs35705950 | TOLLIP rs3750920 | TOLLIP rs111521887 | TOLLIP rs5743894 | G-T-G-C haplotype* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG (n = 30) | GT/TT (n = 67) | P value | CC (n = 19) | CT/TT (n = 78) | P value | CC (n = 60) | CG/GG (n = 37) | P value | TT (n = 59) | TC/CC (n = 38) | P value | No (n = 89) | Yes (n = 8) | P value | |
| BAL features | |||||||||||||||
| Lymphocytes | 13.8 (9.6–21.1) | 11.0 (6–19.2) | 0.117 | 11.1 (5.2–13.7) | 12.0 (7.0–20.2) | 0.322 | 12.2(6.6–19.4) | 11.0 (6.5–20.9) | 0.615 | 12.2(6.6–19.4) | 9.7 (6.0–20.6) | 0.384 | 12(6.6–20.9) | 8.1 (6.4–16.0) | 0.377 |
| Neutrophils | 6.4 (3.4–12.1) | 5.4 (3.2–11.0) | 0.283 | 4.0 (3–8) | 5.8 (3.2–11.2) | 0.329 | 5.0 (3.2–9.9) | 8.0 (3.8–12.2) | 0.142 | 4.9 (3.2–9.4) | 8.0 (4.2–13.5) | 0.082 | 5.5 (3.2–10.5) | 11.2(7.9–18.2) | 0.085 |
| Eosinophils | 3.0 (1.2–12.7) | 2.8 (1.4–5.8) | 0.660 | 3.0 (0.6–9.2) | 2.7 (1.4–2.6) | 0.882 | 2.5 (1.1–5.8) | 4.0 (1.6–9.3) | 0.356 | 2.4 (1.0–5.8) | 4.4 (1.7–9.9 | 0.210 | 2.8 (1.1–6.6) | 4.1 (2.3–10.4) | 0.332 |
| Radiographic features | |||||||||||||||
| UIP + probable UIP | 5 (17.9 %) | 23 (34.8 %) | 0.238 | 4 (22.2 %) | 24 (31.5 %) | 0.743 | 16 (28.1 %) | 12 (32.5 %) | 0.490 | 16 (28.6 %) | 12 (31.6 %) | 0.616 | 25 (29.0 %) | 3 (37.5 %) | 0.755 |
| Indeterminated for UIP | 8 (28.6 %) | 13 (19.7 %) | 5 (27.8 %) | 16 (21.1 %) | 11 (19.3 %) | 10 (27.0 %) | 11 (19.6 %) | 10 (26.3 %) | 20 (23.3 %) | 1 (12.5 %) | |||||
| Alternative diagnosis | 15 (53.6 %) | 30 (45.5 %) | 9 (50 %) | 36 (47.4 %) | 30 (52.6 %) | 15 (40.5 %) | 29 (51.8 %) | 16 (42.1 %) | 41 (47.7 %) | 4 (50.0 %) | |||||
| Honeycombing | 13 (46.4 %) | 24 (36.4 %) | 0.361 | 8 (44.4 %) | 29 (38.2 %) | 0.624 | 25 (43.9 %) | 12 (32.4 %) | 0.268 | 24 (42.9 %) | 13 (34.2 %) | 0.400 | 35 (40.7 %) | 2 (25.0 %) | 0.385 |
| Fibrosis score | 9.5 (7–12) | 8 (5.0–10.8) | 0.023 | 8.5 (5.8–11) | 8 (6–11) | 0.979 | 9 (6.3–11) | 8 (6.0–11.0) | 0.482 | 8.5 (6.0–11.0) | 8 (6.0–10.8) | 0.516 | 8 (6.0–11.0) | 10 (6.0–12.5) | 0.455 |
| Histopathological features | |||||||||||||||
| Honeycombing | 6 (33.3 %) | 14 (38.9 %) | 0.690 | 3 (21.2 %) | 17 (42.5 %) | 0.160 | 12 (33.3 %) | 8 (44.4 %) | 0.425 | 11 (31.4 %) | 9 (47.4 %) | 0.247 | 18 (35.3 %) | 2 (66.7 %) | 0.274 |
| Area of fibrosis (%) | 31.7 (16.2) | 31.4 (13.1) | 0.941 | 30.7 (17.2) | 31.8 (13.0) | 0.806 | 31.1 (14.7) | 32.2 (12.9) | 0.775 | 31.0 (14.9) | 32.4 (12.6) | 0.735 | 31.5 (14.2) | 30.2 (12.4) | 0.866 |
| Fibroblast foci | ´ | ||||||||||||||
| 0 | 10 (55.6 %) | 12 (33.3 %) | 0.270 | 3(21.4 %) | 19 (47.5 %) | 0.206 | 17 (47.2 %) | 5 (27.8 %) | 0.390 | 16 (45.7 %) | 6 (31.5 %) | 0.600 | 20 (39.2 %) | 2 (66.7 %) | 0.561 |
| 1 | 3 (16.7 %) | 7 (19.5 %) | 4 (28.6 %) | 6 (15.0 %) | 6 (16.7 %) | 4 (22.2 %) | 6 (17.2 %) | 4 (21.1 %) | 10 (19.6 %) | 0 (0.0 %) | |||||
| >1 | 5 (27.7 %) | 17 (47.2 %) | 7 (50.0 %) | 15 (37.5 %) | 13 (36.1 %) | 9 (50.0 %) | 13 (37.1 %) | 9 (47.4 %) | 21 (41.2 %) | 1 (33.3 %) | |||||
| Fibrosis distribution | |||||||||||||||
| Peribronchiolar | 6 (33.3 %) | 7 (20.0 %) | 0.677 | 4 (28.6 %) | 9 (23.1 %) | 0.816 | 10 (28.6 %) | 3 (16.7 %) | 0.428 | 10 (29.4 %) | 3 (15.8 %) | 0.438 | 12 (24.0 %) | 1 (33.3 %) | 0.849 |
| Peribronchiolar (+paraseptal or subpleural) | 8 (44.4 %) | 20 (57.1 %) | 7 (50.0 %) | 21 (53.8 %) | 16 (45.7 %) | 12 (66.7 %) | 16 (47.1 %) | 12 (63.2 %) | 27 (54.0 %) | 1 (33.3 %) | |||||
| Heterogeneous | 3 (16.7 %) | 7 (20.0 %) | 2 (14.3 %) | 8 (20.5 %) | 7 (20.0 %) | 3 (16.7 %) | 6 (17.6 %) | 4 (21.1 %) | 9 (18.0 %) | 1 (33.3 %) | |||||
| Undetermined | 1 (5.6 %) | 1 (2.9 %) | 1 (7.1 %) | 1 (2.6 %) | 2 (5.7 %) | 0 (0.0 %) | 2 (5.9 %) | 0 (0.0 %) | 2 (4.0 %) | 0 (0.0 %) | |||||
| Inflammatory infiltrate | |||||||||||||||
| Light | 14 (77.8 %) | 24 (66.7 %) | 0.532- | 10 (71.4 %) | 28 (70.0 %) | 0.920 | 26 (72.2 %) | 12 (66.7 %) | 0.756 | 26 (74.3 %) | 12 (63.2 %) | 0.534 | 37 (72.5 %) | 1 (33.3 %) | 0.148 |
| Moderate-severe | 4 (22.2 %) | 12 (33.3 %) | 4 (28.6 %) | 12 (30 %) | 10 (27.8 %) | 6 (33.3 %) | 9 (25.7 %) | 7 (36.8 %) | 14 (27.5 %) | 2 (66.7 %) | |||||
| Peribronchiolar metaplasia | 2(1–3) | 1 (1.9–2.3) | 0.749 | 1 (0.0–2.8) | 2(1.0–3.0) | 0.390 | 2(0.8–3.0) | 1 (1.0–2.0) | 0.829 | 2(0.5–3.0) | 1 (1.0–2.5) | 0.623 | 2 (1.0–3.0) | 1 (1–1) | 0.353 |
| Giant cells granulomas present | 10 (55.5 %) | 6 (16.7 %) | 0.003 | 6 (42.9 %) | 10 (25.0 %) | 0.208 | 10 (27.8 %) | 6 (33.3 %) | 0.673 | 10 (28.6 %) | 6 (31.6 %) | 0.817 | 15 (29.4 %) | 1 (33.3 %) | 0.885 |
Values are presented mean (SD) or as median (Q1-Q3) and counts as n (%).
BAL sample analyses revealed a tendency for a larger amount of neutrophils in patients bearing at least one of the minor alleles of T887 (G) and T894 (C) polymorphisms. Conversely, for MUC5B rs35705950 this trend was noted but for the ancestral allele (G). Accordingly, carriers of the G-T-G-C haplotype, which is associated with poorer prognosis, showed a median neutrophil amount that more than doubled that of non-carriers although not reaching statistical significance (P = 0.085) (Table 5).
No additional significant associations emerged between the probed SNPs or respective haplotypes and histopathological features of fibrosis (Table 5).
DiscussionThis study investigated specific genetic variants as determinants of susceptibility and survival in patients with fHP. The MUC5B promoter rs35705950 variant and TOLLIP rs3750920, rs111521887 and rs5743894 variants are associated with susceptibility to fHP in this Portuguese cohort. Although such an association had been previously reported for MUC5B rs35705950,20-22 this is the first study implicating TOLLIP variants as susceptibility determinants in fHP. Notably, we also found that a haplotype defined by these four variants is significantly associated with poorer survival in these patients.
The role of the MUC5B polymorphism in the development and progression of lung fibrosis has been extensively investigated in IPF and is considered the most influential genetic risk factor of this disease.13-15,30 The involvement of MUC5B in HP was described in recent studies20-22 with results corroborated by those presented herein. Furusawa et al identified six common IPF genetic risk variants with a positive direction of risk effect in HP cases, with the MUC5B variant showing the strongest association.21
Other common variants reportedly associated with IPF susceptibility include SNPs in the genomic region of the TOLLIP gene. Notwithstanding some inconsistent reports, it is generally accepted that TOLLIP polymorphisms play a role in IPF pathophysiology. The minor alleles of intronic SNPs rs111521887 and rs5743894 were reported to confer susceptibility to IPF, while that of the rs5743890 SNP was found to reduce such risk.30,31 Moreover, TOLLIP’s exonic variant rs3750920 was associated with a favorable response to N-acetylcysteine, with carriers of the TT genotype showing a significantly reduced composite endpoint risk, including death, hospitalization, and FVC decline.17
Having probed five TOLLIP SNPs, including those above, we have uncovered a significant correlation between genotypes of TOLLIP rs3750920, rs111521887 and rs5743894 and susceptibility to fHP, with the minor alleles associating with higher risk. Interestingly, none of these SNPs independently associated with IPF risk in a previous study with Portuguese patients,18 raising the possibility that unlike MUC5B rs35705950, TOLLIP can be an informative marker in a future strategy for differential diagnosis between the two fibrotic diseases, at least in the Portuguese. Several studies have implicated TOLLIP as playing a protective role in autophagy and apoptosis.32,33TOLLIP’s expression is decreased in IPF lungs when compared to normal lungs,32 which has been linked to minor allele carriers of variants T887, T894 and T890, further suggesting that these could modify the course of disease.30 In our study, the minor alleles of the former two variants tended to or associated with susceptibility (respectively) and integrated the haplotype associated with poorer prognosis in fHP. Interestingly, genotypes containing at least one minor allele of TOLLIP variants rs111521887 and rs5743894 were significantly associated with lower baseline FVC% pred, whereas for rs3750920 the decrease was of borderline statistical significance. In Japanese fHP patients genotyped for TOLLIP rs3750920 and rs5743899 (residing between T887 and T894 and not evaluated here), Katayanagi et al reported that the GG (minor allele) genotype of rs5743899 only, was associated with rapid FVC deterioration over time.34 The relationship between rs3750920 genotypes and FVC at baseline was not stated.
The lack of association between TOLLIP rs5743890 and fHP risk is in line with that reported by Ley et al.20 We are not aware of studies concerning TOLLIP rs5743854 in the context of fHP.
We found no association between the MUC5B variant and baseline FVC, in contrast with a recent study in Polish patients.35 However, the latter reported a MUC5B MAF of only 17 %, with a GT/TT prevalence of 30.2 %, which contrasts strongly with those reported here and elsewhere.20
Ley et al reported that the MUC5B rs35705950 minor allele was associated with radiographic evidence of moderate to severe fibrosis and traction bronchiectasis in HP patients.20 In our study the minor allele associated with a lower total score of fibrosis; nonetheless, HRCT fibrosis was scored differently and the median scores in both genotypic groups (GT/TT=8 and GG=9.5) already represent at least moderate fibrosis.
The use of SNP haplotypes within regions of interest previously identified by single-SNP genome-wide association studies can prove very useful in uncovering and fine-mapping genetic associations,36 and provide better insights into the influence of genetic loci than the study of SNPs individually.37,38 We show that four haplotypes, defined by the MUC5B promoter SNP and the three TOLLIP SNPs that associated individually with susceptibility to fHP, were also strong predictors of fHP risk. Remarkably, carriers of the haplotype G-T-G-C had a significant decrease in survival compared to non-carriers, with the median survival time differing by a remarkable 21 months. This association was confirmed by multivariate analysis adjusting for age and lung function. We had previously reported an association of MUC5B plus TOLLIP-defined haplotypes with susceptibility and survival in IPF using a similar approach,18 suggesting that the region comprising these variants contains or segregates with an as-yet unidentified genetic determinant(s) that influence(s) the clinical course of both disorders. This, in spite of the absence of an independent association of each TOLLIP variant and the MUC5B promoter SNP with fHP survival. These results are in accordance with the report by Ley et al, who also found no independent association between MUC5B rs35705950 or TOLLIP rs5743890 and survival in patients with fHP.20
The G-T-G-C haplotype is also associated with a feature that can negatively impact in disease prognosis. All carriers of the haplotype had been subjected to bird exposure, an antigen that is frequently described in more severe and fibrotic forms of the disease,39 despite the lack of an association with compromised lung function in our study.
Overall, our findings suggest that TOLLIP variants, as well as haplotypes (incorporating MUC5B rs35705950), could be of potential use for patients’ risk stratification and as markers of prognostic value in fHP. Multicenter and prospective studies are needed to validate these results and to elucidate the role of both genes in the pathophysiology of hypersensitivity pneumonitis.
FundingThis work was supported by the Portuguese Society of Pulmonology (SPP) (SPP/Bolsa Novartis 2012 grant); Boehringer-Ingelheim (Unrestricted Research Grant); Fundação para a Ciência e a Tecnologia, I.P. (PTDC/MEC-RES/0158/2020 grant); and Fundação Amélia de Mello and D. José de Mello.
JCR is funded by Fundação para a Ciência e a Tecnologia through the Stimulus for Scientific Employment Individual Support (2020.01350.CEECIND).
We are indebted to Instituto de Saúde Pública da Universidade do Porto (ISPUP) for providing samples from the EPIPorto cohort to be used as controls; we thank the cohort management and all participants. We are grateful to Catarina Teixeira Antunes and Joana Filipa Amorim Reis for assistance in the genotyping.